Abstract
Introduction
Diffuse large B-cell lymphoma (DLBCL) represents the most frequent NHL and comprises two molecular subgroups, namely germinal center B-cell-like (GCB) and activated B-cell-like (ABC) subgroups, each having distinct gene expression and genetic features. Follicular lymphoma (FL) is an indolent cancer that shares genetic features with DLBCL, particularly the GCB subgroup. Despite a growing knowledge about molecular features of these cancers. There remains a limited understanding of the recurrence of noncoding mutations that may impact gene function or expression in DLBCL, owing, in part, to their genetic complexity and a paucity of whole genome sequence (WGS) data. We sought to uncover novel mutation patterns through global meta-analysis of a large cohort of DLBCLs comprising 353 discovery cases, an independent set of 300 validation cases and comparison to genomes from 124 FLs.
Methods
DLBCL exomes with matched constitutional DNA (n=200) from multiple studies and genomes from prior publications (40), ICGC (41) and 72 additional patients (n=153) were analyzed using a unified somatic single nucleotide variant (SNV) pipeline. Significantly mutated genes (SMGs) were identified using OncodriveFML. Recurrent regions affected by noncoding mutations ("peaks") were determined using a novel approach - an extension of the rainfall plot computation followed by wavelet analysis. Structural variants (SV) were detected using Manta and copy number variations (CNVs) were determined using read-depth strategies. A separate set of 124 FL genomes was analyzed similarly. RNA-seq data were available for 143 of the DLBCL genomes and were processed using Salmon and analyzed for differential expression using DeSeq2. Using a custom capture panel, 82 genes were sequenced in some of the discovery cases and an independent DLBCL cohort comprising 300 cases. This allowed us to confirm and determine the recurrence of mutations and SVs affecting coding exons and UTRs. RNA-seq and SNP6.0 arrays were also applied to all validation cases to confirm patterns of coordinated expression.
Results
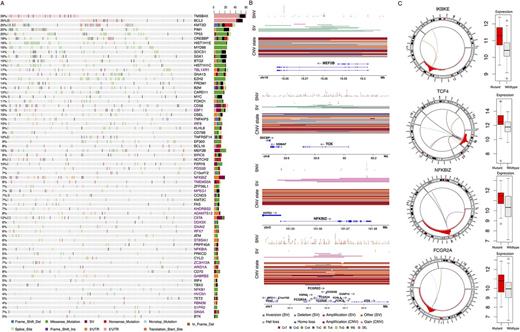
We detected 52 SMGs (Fig. 1A) and 4382 mutation peaks among the DLBCL meta-cohort. Up to 24 of these have not previously been ascribed to DLBCL (Fig. 1A, purple). Most mutation peaks proximal to genes affected the first intron or promoter, consistent with AID activity. Many of the affected genes are known targets of aberrant somatic hypermutation though some novel targets of this process were identified. Non-genic peaks tended to affect putative distal enhancers of BCL6, IRF4 and PAX5. A subset of the noncoding regions and novel SMGs exhibited specificity for mutation within either ABC or GCB DLBCLs, including some genes involved in NF-kB signalling. Less than 10% of peaks were mutated among the FLs and these were largely those with a higher mutation burden in GCB DLBCL. We identified numerous genes recurrently affected by SVs, SNVs and CNVs and determined the subset of those with expression altered in the presence of such candidate regulatory mutations. For example, MEF2B and TOX both showed patterns indicative of tumour suppressor function, including a large number of focal deletions (Fig. 1C,D). In contrast, we observed patterns consistent with a role of mutations promoting the expression of each of TCF4, ΙΚΒΚΕ, NFKBIZ (two regulators of NF-κB) and the Fc γ receptor locus (Fig. 1B). TCF4 harbored mainly SV breakpoints that may disrupt some of the many annotated isoforms and leave others intact whereas NFKBIZ was predominantly affected by mutations in the 3'UTR. The FC γ locus contained numerous examples of focal gains and rearrangements involving this region and these were also detected in some of the FLs.
Conclusion
This integrative genomic analysis offers numerous insights into the biology of DLBCL and features that distinguish it from indolent cancers arising in the germinal center. The novel coding and noncoding mutations found herein and those enriched in either DLBCL subgroup will aid to extend our understanding of the molecular underpinnings of these and their relationship to FL. Additional genes involved in NF-κB have been found to be deregulated through unique genetic mechanisms that have evaded detection though exome-based analyses. Further, the expression and sequence of Fc γ receptors is thought to impact the efficacy of anti-CD20 therapies, though the prevalence of somatic alterations has not been thoroughly explored.
Sehn: Celgene: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; Janssen: Consultancy, Honoraria; Abbvie: Consultancy, Honoraria; Roche/Genentech: Consultancy, Honoraria; Seattle Genetics: Consultancy, Honoraria. Steidl: Affimed Therapeutics: Consultancy; Juno Therapeutics: Consultancy. Connors: F Hoffmann-La Roche: Research Funding; NanoString Technologies, Amgen, Bayer, BMS, Cephalon, Roche, Genentech, Janssen, Lilly, Merck, Seattle Genetics, Takeda,: Research Funding; NanoString Technologies: Research Funding; Lilly: Research Funding; Janssen: Research Funding; Genentech: Research Funding; Cephalon: Research Funding; Bristol-Myers Squibb: Research Funding; Bayer Healthcare: Research Funding; Seattle Genetics: Research Funding; Merck: Research Funding; Amgen: Research Funding; Takeda: Research Funding. Scott: Janssen: Consultancy, Honoraria; Celgene: Consultancy, Honoraria; BCCA: Patents & Royalties: Patent describing molecular subtyping of DLBCL licensed to NanoString Technologies. Patent describing measurement of the proliferation signature in MCL.. Morin: Epizyme, Inc: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal